Documentation
GenomicShiny-Cov was tested in windows 10 and Debian/Ubuntu distros, you must be installed R v.4.0 or higher, is not necessary to install Rstudio to run GenomicShiny-Cov.
System requirements
Ubuntu 20.04 / 21.10 / 22.04
Debian 10
Install dependencies for linux ditros
Install dependencies by aptitude:
libprotobuf-dev
libjq-dev
protobuf-compiler
libudunits2-dev
libgdal-dev
Instalation
To run locally the application must be installed first shiny package:
### shiny installation
install.packages("shiny")
Runing from Github
Copy the following link to install and run GenomicShiny-Cov:
### Copy the link and run in R console
shiny::runGitHub("GenomicShiny-Cov","FranciscoAscue",
launch.browser = TRUE)
Clone the repository
Linux To run locally clone the repository:
git clone https://github.com/FranciscoAscue/GenomicShiny-Cov.git
cd GenomicShiny-Cov
Rscript app.R
Windows
Download the code from the following LINK and unzipped Open the project in R console :
runApp(launch.browser = TRUE)
Build docker image
git clone https://github.com/FranciscoAscue/GenomicShiny-Cov.git
cd GenomicShiny-Cov
docker build .
Pull docker images from DockerHub
docker pull fascue/genomicshiny0.1
Input Files and Usage
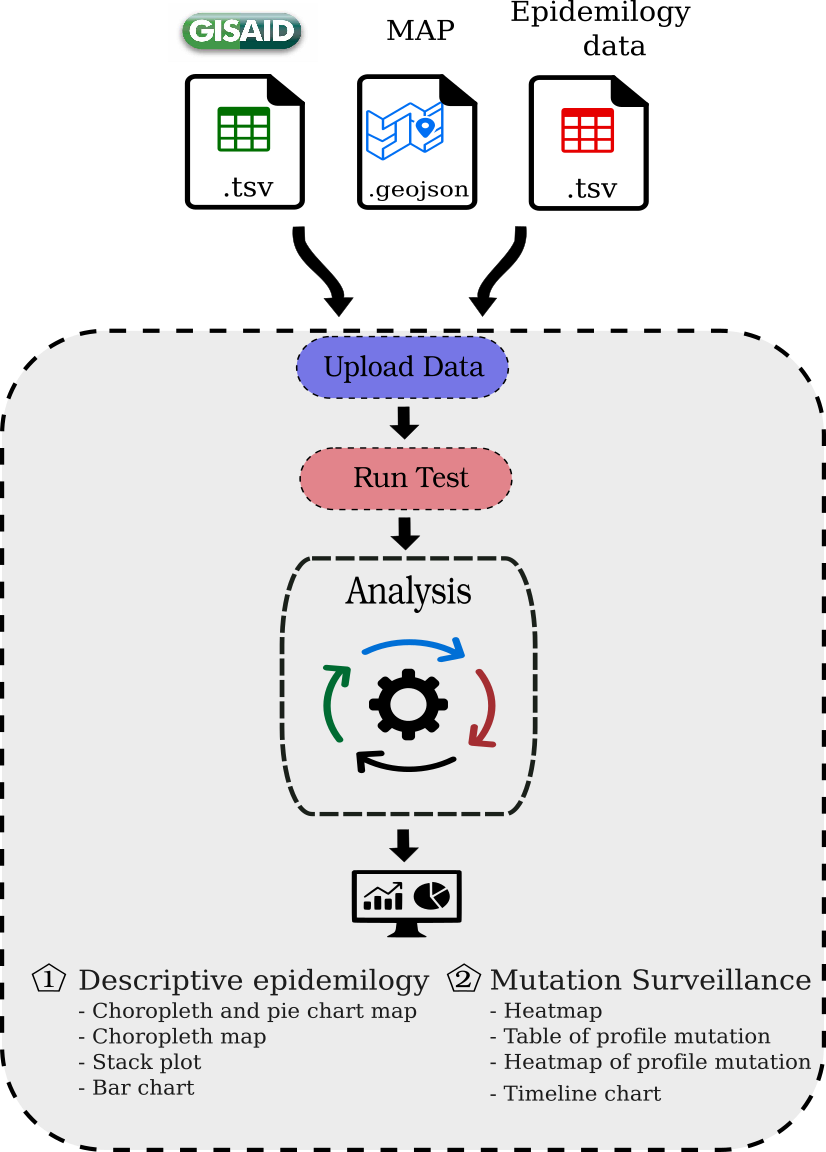
| File | Description | i.e. Peru | i.e. USA | i.e brazil | i.e. Colombia |
|---|---|---|---|---|---|
| Geojson map | Vectorial map | GeoJSON | GeoJSON | GeoJSON | GeoJSON |
| Metadata from GISAID | Pacient status metadata from EpiCoV GISAID | peru.tsv | usa.tsv | brazil.tsv | colombia.tsv |
| Epidemiology data | Positive or death cases reported | epiPeru.csv | epiUsa.tsv | epiBrazil.csv | epiColombia.csv |